
Unilever Centre for Molecular Sciences Informatics
Department of Chemistry
University of Cambridge
Cambridge CB2 1EW, UK
Open Data (OD) is an emerging term in the process of defining how scientific data may be published and re-used without price or permission barriers. Scientists generally see published data as belonging to the scientific community, but many publishers claim copyright over data and will not allow its re-use without permission. This is a major impediment to the progress of scholarship in the digital age. This article reviews the need for Open Data, shows examples of why Open Data are valuable and summarises some early initiatives in formalising the right of access to and re-use of scientific data.
I am grateful to the editor for her invitation to contribute an article on "Open Data" in this issue updating Open Access since the last special issue on the topic. The term "Open Data" (OD) is realtively new and may be unfamiliar to many readers. It springs from some of the same roots as "Open Source" and "Open Access" but it is dangerous to try to make easy extrapolations from these. Although not yet common, OD is used an a variety of domains, several of which are outside the scope of this article, which is limited to the publication of scientific results and discourse. Moreover there has been some very recent activity in starting to define OD more precisely and it is likely to develop considerably in the next few years.
Since the technology of publishing is changing rapidly I have taken the opportunity of casting this article in a modern form, the "datument". This is a compound document where all the compounds (data, text, software,images, links) are part of the whole. (I and others have repeatedly shown that PDF is a major cause of destruction of scientific information and that datuments - XHTML+XML - are an excellent medium for communicating data). Since the topic of this paper is "Open Data" and since datuments are a key part of the technology that Open Data exposes then I am using the medium as the message.
For that reason there are hyperlinks in the text, but not conventional references. There are a number of active objects which should work in all modern browsers. You may find it useful to try more than one.
The terms "Open" and "Open Access" are used in very variable and confusing ways. In this article the sense is similar to "Open" in "Open Source" [software] where the term "Free" (or libre) is also used. The following definition of Open Source (libre) software uses phrases that map fairly well onto our use of "Open Data"...
[...] that can be used, studied, and modified without restriction, and which can be copied and redistributed in modified or unmodified form either without restriction, or with restrictions only to ensure that further recipients can also do these things. [...] may be either accompanied by a software license saying that the copyright holder permits these acts (a free [...] licence), or be released into public domain, so that these rights automatically hold. 1
English does not have a common term for "libre" and so "free" ("as in speech, not as in beer" 2) cannot specify intent. Although "Open" in software normally means libre, there is an increasing (and unfortunate) movement towards using "Open Access" to mean gratis and not libre. In this article "Open Data" refers to libre consistently.
An important concept of Open Data is "re-use". This is developed below, and represents the use of the data, normally without explicit permission, for studies foreseen or not foreseen by the original creator. These include aggregation (into databases), parameters in simulations, and "mashups" where data from different sources are combined to give new insights.
A common attribute of Open Data is "removal of permission barriers" (explained below) and to avoid repeating this phrase too often I have sometimes shortened it to "permissionFree".
The term "Open Access publisher" is ungainly but useful and refers to a publisher who labels some or all of their products "Open Access".
The term "Community Norms" represents an acceptance of appropriate behaviour which has moral, but no legal force.
This article is split into the following sections: an introduction to the topic; an in-depth analysis of an example of the need for OD (taken from chemistry); and a summary of recent developments and recommendations. To show the power and need for OD, some of the material in the second section is interactive and I am grateful for the editor in allowing me to submit the manuscript in XHTML
The examples in this article are from chemistry (but require no specific chemical knowledge). It is recognised that practice in publishing and re-using data varies greatly between disciplines. Some, like bioscience, have a long tradition of requiring data to be published and then aggregated in publicly funded databanks. Others in "large science" have a well-developed data re-use policy and require data from telescopes, satellites, particle accelerators, neutron sources, etc. to be made universally available for re-use. In these areas Norms are often sufficient for the practice of Open Data. In "small science" by contrast the unit of research is the lab or individual ("small" does not reflect the importance of the diciplien which may be numerically very large). These disciplines typically result in many independent publications which report individual experiments (I coined the neologism "hypopublication" to express the disjointed nature of this information).
This article concentrates on the role of the scholarly publication process. There are other ways in which data are made available, and where the precept of OD will be important. This article, however, concentrates on balance between the scholarly publisher, the author and the reader.
I first realised the Open Data problem 5 years ago when Henry Rzepa and I submitted a manuscript to the Journal of Chemical Information and Modeling, published by the American Chemical Society. As discussed in depth later, many scientific manuscripts consist of a "full-text" manuscript and additional "supplemental data" or "supporting information" (SI) (the term varies with publisher and I shall use SI hereafter. In many cases the role of SI is to provide information which is too large or "boring" to fit in the full-text but which is necessary for a reader who wants to be sure that the experiment has been carried out correctly and that the right conclusions have been drawn. One ideal is that it could be a copy of the original lab notebook (and some innovators such as Jean-Claude Bradley are deliberately publishing full details on the web). This level of detail is rare and it is normally more compact, but some SI can run to 200 pages of spectra and analytical data.
Although we had (reluctantly) agreed to transfer of copyright (TrOC) of the fulltext to the ACS (there are very few OA journals in chemistry) we wished to retain rights over the SI. However the ACS stated (and still states 2007-12):
Electronic Supporting Information files are available without a subscription to ACS Web Editions. All files are copyrighted by the American Chemical Society. Files may be downloaded for personal use; users are not permitted to reproduce, republish, redistribute, or resell any Supporting Information, either in whole or in part, in either machine-readable form or any other form. For permission to reproduce this material, contact the ACS Copyright Office by e-mail at copyright@acs.org or by fax at 202-776-8112.
Rzepa and I were concerned about this, especially since most of the SI files pubshed in chemistry are simply a collection of facts (see section (b) for details). In almost all cases these are a record of the experiment and include temperatures, materials, analytical results, etc. or simple copy of the computer output of a simulation. We assumed that this notice was an oversight since facts cannot be copyrighted. However after repeated correspondence the ACS made it clear that this was deliberate policy and would not be waived except in special cases. (We believe that we are the only authors for which this has been waived).
We believe that the data in SI are extremely valuable for re-use. A classic example of re-use is Mendeleev's use of published data to propose the Periodic Table of the Chemical Elements. In this case the published data (melting points, colours, densities, etc.) were often not collected for a specific purpose other than the worthy belief that data was, per se, valuable. This is still as true today; what scientific quantity could possibly be deduced from ancient Chinese eclipse records? Yet K D Pang, K Yau and H-H Chou showed that this gave a value for the variation of the earth's rotation during the postglacial rebound and from this deduced a value for the lower mantle viscosity of the earth. There are many examples of scientific effects hidden in routine data (for example the discovery of pulsars) and I assert axiomatically that data, per se, is valuable for re-use.
It is important to realise that SI is almost always completely produced by the original authors and, in many cases, is a direct output from a computer. The reviewers may use the data for assessing the validity of the science in the publication but I know of no cases where an editor has required the editing of SI. The editorial process, adds nothing to the content and adds no value other than publishing it alongside the full-text (which may have had valuable input from the reviewers and editors). It is probable that many publishers will not have the technical expertise to evaluate the validity of the SI by themselves.
It is clear, however, that the publishers such as ACS and Wiley assert control over this information, the latter through an explicit notice () embedded in the SI. To avoid copyright problems in quoting this I have transcribed it as:
"Angewandte Chemie [logo omitted] Supporting Information for Angew Chem Int Ed. Z52910 © Wiley-VCH 2003 69451 Weinheim, Germany"
Wiley defends its copyright aggressively as Shelley Batts, a PhD student discovered when she challenged science reported in a paper published in a Wiley journal. She copied a graph from the paper and mounted it on her website, and later received the following letter from Wiley (When Fair Use Isn't Fair):
Re: Antioxidants in Berries Increased by Ethanol (but Are Daiquiris Healthy?) by Shelly Bats [sic, PMR]
http://scienceblogs.com/retrospectacle/2007/04/antioxidants_in_berries_increa.php
The above article contains copyrighted material in the form of a table and graphs taken from a recently published paper in the Journal of the Science of Food and Agriculture. If these figures are not removed immediately, lawyers from John Wiley & Sons will contact you with further action.
There was much discussion on the blogosphere and later some suggestion from Wiley that this was overzealous on their part. In the end Batts retyped the data from the original article (fully legal) and redrew the graphs using her own software (perfectly legal). But what a supreme waste of human effort with the added likelihood that there could be transcription errors. However this type of automatic reaction from publishers is not uncommon and shows a corporate mentality of owning all artifacts in publications. The key point was that the material in question was numerical facts, presented in graphical form simply because this is a natural way that scientists communicate. Indeed it would be common for a newcomer to science to be told to present their data in graphical or tabular form.
Is this an object worthy of copyright protection? I suspect this has rarely been tested in court in the case of scientific data. It is universal that no scientist intends or expects to receive any monetary remuneration for a scientific article. They are expected to publish the supporting facts. In almost all cases the actual presentation of those facts is dictated by the needs of the science and not by creative works of the publisher.
I felt strongly that data of this sort should by right belong to the community and not to the publisher and started to draw attention to the problem. I first looked at the basis of Open Access, and particularly the declarations from Berlin, Budapest and Bethesda ("BBB"). The phrase that speaks most closely to Open Data comes from the Budapest version:
... By "open access" to this literature, we mean its free availability on the public internet, permitting any users to read, download, copy, distribute, print, search, or link to the full texts of these articles, crawl them for indexing, pass them as data to software, or use them for any other lawful purpose, without financial, legal, or technical barriers other than those inseparable from gaining access to the internet itself.
This is admirably clear and, I assumed, would be universally understood. An Open Access document, or journal, carried the explicit requirement that the whole contents - text, data, metadata - could be re-used for whatever purpose without further explicit permission. Peter Suber has repeatedly described the financial barriers (mainly journal subscriptions, but also fees for re-use of material) as "price barriers" and legal barriers (copyright, publisher contract, etc.) and technical barrierss (logins, limited visibility, limited durations, etc.) as "permission barriers" and I shall use these terms hereafter. In general this article is concerned with permission barriers.
It seemed axiomatic that "Open Access publishers" should make it explicit that there were no permission barriers by announcing this on their websites, and many do. The most convenient way is to attach a licence to the content, and the Creative Commons - Acknowledgement (CC-BY) fulfills the requirements and is the most common. However several sites had "Open Access" labels but restricted the use of the content, most commonly to "non-commercial" (e.g. through the Creative Common non-Commercial (CC-NC) licence). (In passing we note that the Creative Commons licences were developed for creative works in arts, music, literature etc, and are not ideally suited to scholarly publications and even less so for scientific data, e.g. the use of the word "sampling" in CC-NC. Nevertheless CC-BY is an excellent first step in allowing authors to make their data available for re-use, not least because it acts as a statement of moral intent which would be universally accepted by the community (except those publishers requiring TrOC)).
Since "Open Data" was a rare term at this stage I initiated 2 activities:
More recently I also started a blog and have found that campaigning for Open Data has become one of the main themes. At times I have reported practice in Open Access and specifically permissions barriers. I have also campaigned by writing to publishers for information - sadly many of them to fail to acknowledge my requests (discussed below).
Recently there has been a convergence of two communities in the use of "Open Data". The web community is concerned with access to data on the web including the rights of individuals to retrieve their data from companies to which they have submitted it. In 2007 Paul Miller of Talis ran a session at WWW2007 on "Open Data" and Edd Dumbill has a recurrent theme of "Open Data" at the XTech (XML) meetings. As a result I suggested on the Wikipedia page that OD has been used to represent:
There I also suggested arguments commonly made on behalf of Open Data:
Along with other protagonists I was invited to address the Scientific technical Medical publishers in 2005 on "Open Data" and found considerable support for the concept. The ALPSP and STM issued the following statement in 2006:
Publishers recognise that in many disciplines data itself, in various forms, is now a key output of research. Data searching and mining tools permit increasingly sophisticated use of raw data. Of course, journal articles provide one ‘view’ of the significance and interpretation of that data – and conference presentations and informal exchanges may provide other ‘views’ – but data itself is an increasingly important community resource. Science is best advanced by allowing as many scientists as possible to have access to as much prior data as possible; this avoids costly repetition of work, and allows creative new integration and reworking of existing data.
and
We believe that, as a general principle, data sets, the raw data outputs of research, and sets or sub-sets of that data which are submitted with a paper to a journal, should wherever possible be made freely accessible to other scholars. We believe that the best practice for scholarly journal publishers is to separate supporting data from the article itself, and not to require any transfer of or ownership in such data or data sets as a condition of publication of the article in question.
This again is admirably clear. If this advice were followed then I might not be writing this article. Sadly the STM's exhortations are not followed by all their members.
It is also important to note that published scientific information per se can be of considerable commercial value. While bioscience data is aggregated and made completely freely available, chemical information (abstracts, patents, physical data) is aggregated and resold. I have informally estimated that this market is worth several billion USD with Chemical Abstracts (ACS), Beilstein (Elsevier) and Wiley major suppliers of databases and similar products. There is therefore a conflict between the role of s scholarly publisher in making data available and the commercial databases supplier to whom literature data is a revenue stream. This was epitomised by the action of the ACS in trying to drastically restrict the activities of the government-funded Pubchem collection of compounds of biological interest (reported in Nature).
In this section we show in detail what Open Data are and how they can be valuably re-used. The exemplars are from chemistry, but the reader should require no specialist knowledge. There are two sections. The first explores fulltext and SI from Open Access CC-BY publications. It is worth noting in passing that it would have been virtually impossible to write this using similar material from Elsevier or other closed access publishers. The multiple re-use of material could have cost several hundred USD, the requests for permission could have taken months (see Nico Adams' blog) and would probably have been forbidden in some cases). As it is I have appended all material as supplemental data to this publication, which is allowed by the original authors and publishers. without having to ask permission. The second shows how very large amounts of data can be automatically extracted from SI in published papers as long as the SI is openly re-usable.
NOTE: This section uses the power of HTML to hyperlink material. The reader should be already viewing this section in an HTML browser and should be prepared to follow hyperlinks as indicated.
I have taken three recent papers from Beilstein Journal of Organic Chemistry chosen to illustrate the components:
(I pass no judgment on the science other that to reassure readers that this is a fully peer-reviewed journal). Like many other journals most of the material is offered in HTML and PDF, the exemption being the SI (here called "Additional material"). This can be in many forms, the commonest being *.doc (Word), PDF, image formats (PNG, JPG, etc.), movie formats and chemistry such as CIF, CML, (Chemical Markup Language) etc. .
The SI is not normally part of the PDF/HTML full-text and is in one or more separate files, usually with hyperlinks from the fulltext (in the HTML version it is called "Additional Files"). The PDF "fulltext" of a paper normally concentrates on the visual aspect of a paper, while the HTML preserves much of the structure and allows multiple click-throughs and interactive dataTypes.
At this stage the reader should load [localcopy of PDF]. The first page contains the section "Background" with this excerpt:
| ... Many of these reactions revealhigh diastereoselectivities, which has been exploited to access a range ofnatural products and their derivatives.[8,9] An example of Casiraghi's fromthe castanospermine repertoire showing the numbering system used in thisarticle is shown in Scheme 1.[4] |
 |
| Scheme 1 |
| An example of a vinylogousMukaiyama aldol in a natural product synthesis. Reagents and conditions: a)RCHO 2, SnCl4 (1.2 equiv), ether, -85°C. |
The key point is that Scheme 1 above is chemical data. These are conventional representations of chemical structures - the lower bold numbers are used to identify the compounds, the smaller upper numbers to identify the atoms. The arrow represent a chemical reaction. The scheme is the natural and universal way of conveying the factual information about what a chemical is. It is not a work of art, although different tools create images with different styles. It is essential to the understanding of the paper.
Under conventional practice this could be regarded by a publisher as a creative work and therefore requiring explicit permission for re-use. My argument is that this is factual information for which no permission should be required or sought.
The facts in Scheme1 are also valuable for re-use. Although the image is a poor quality GIF it takes a minute to for me to use standard software and create three-dimensional molecules (here shown for compound 3)
The same argument holds for the factual data embedded in the running text and in the images. Here are some excerpts which all chemists would regard as facts:
Synthesis of Adduct 16 from adduct 6a. Reagents and conditions: a) CAN, aq CH3CN, -20°C to rt, 5 h, 82%; b) Br2, MeOH, -20°C, 30 mins, 91%; c) Zn, aq NH4Cl, THF, MeOH, RT, 30 mins, 90%; d) (Boc)2O (4 equiv), THF, DMAP (cat), rt, 18 h, 90%; e) DIBAL-H, THF, -78°C to -20°C, 2 h, 86%; f) Et3SiH, BF3·OEt2, DCM, -70°C, 1 h, 91%; g) TBAF, THF, 10°C, 5 days, 84%.
 |
Figure 4. X-ray structure of 16. |
In paper30 [localcopy PDF] we find diagrams such as:
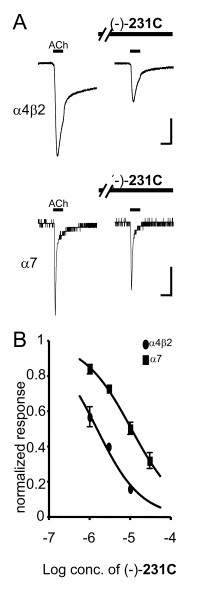 |
"A" is almost certainly produced directly by machine, other than the annotations. A reader might wish to compare the heights of the peaks.Ideally the data should be in numeric form so that they could be automatically analysed by machine; unfortunately in this case the value will probably have to be measured of this graph with a ruler, leading to sizeable errors. "B" is a domain-standard way of presenting dose-response data in biological experiments. Again this is factual data. A reader will probably wish to determine the horizontal difference between the curves ("about 1 log unit"). In passing I note that the use of graphs is often determined by publisher policy, many of whom do not encourage or permit the deposition of SI. In this way the publication process itself leads to very serious loss of factual data.
We now pass to the SI which is shown for paper49. This has 42 pages (not untypical) and consists of factual data on the preparation of all the compounds in the paper together with their analytical data including spectra. (Again it is unfortunately that the data are not in numerical form). The primary purpose of these factual data are to validate the synthesis of the compound - at a simple level every chemical compound has a different spectrum ("fingerprint"). But these data are also extremely valuable for re-use as we shall now show:
Since many scientific facts are still published in textual form it has become essential to have software ("text-mining") that can understand them in this form. This section shows how this can be done, and although it is equally applicable to full-text and SI, we are often explicitly forbidden (in the subscription contract) to text-mine full-text. The following text occurs in the SI of paper 38.
4-Methoxy-3-pyrrolin-2-one-1-carboxylic acid benzyl ester 4d. To a solution of pyrrolinone 4 (0.80 g, 7.08 mmol) in dry tetrahydrofuran (50 ml) at 0C, was added sodium hydride (60% in oil, 0.38 g, 21.2 mmol, 3 eq) and the reaction mixture stirred at 0C for 10 minutes. Benzyl chloroformate (1.1 ml, 7.8 mmol, 1.1 eq) was added at 0C and the reaction mixture stirred at room temperature for 3h. The reaction mixture was quenched with aqueous ammonium chloride (100 ml) and extracted with ethyl acetate (3 x 150 ml). The combined organic layer was dried over magnesium sulfate and concentrated under reduced pressure to give a residue (2.92 g), which was chromatographed on silica-gel (70 g) using ethyl acetate / petroleum ether (7 : 3) as eluant to give 4d (1.1 g, 3.54 mmol, 50%). M.p. 128-129C (from ethyl acetate); (Found: C, 63.15; H, 5.00; N; 5.61%. C13H13NO4 requires C, 63.15; H, 5.30; N, 5.66%); (Found [M+H]+ 248.0922, C13H13NO4 requires 248.0922.); max/cm-1 3056 (C-H arom), 2986, 2943, 2898, 2870, 2856 (C-H aliph), 1781 (O-C=O), 1735 (N-C=O), 1633 (C=C), 1331; H (CDCl3, 300 MHz) 3.83 (3H, s, OCH3), 4.23 (2H, s, H-5), 5.11 (1H, s, H-4), 5.29 (2H, s, Bn), 7.30-7.50 (5H, m, Ar); C (CDCl3, 100.6 MHz) 49.1 (C-5), 58.6 (OCH3), 67.7 (Bn), 94.7 (C-3), 128.1, 128.3, 128.5, 135.5 (Ar), 150.6 (COCarb), 168.5 (C-4), 174.9 (COlact).
This may appear impossible to understand but it has a fairly regular microstructure and our OSCAR toolkits can make a great deal of sense of it. OSCAR-DATA was originally developed (Experimental data checker: better information for organic chemists) through sponsorship of the Royal Society of Chemistry (Experimental Data Checker Homepage) and is now developed as a standalone data-checker. Simply cutting and pasting the above text into OSCAR-DATA gives the result:
 |
It can be seen that OSCAR has "understood" the numeric parts and these are now in semantic form; OSCAR can reconstruct what the spectrum would have looked like if the authors had published it:
OSCAR3 can similarly
"understand" the chemical language in the text; here is a simple example from
paper30.
Here part of the fulltext has been pasted straight into OSCAR3, which has automatically recognized all words and phrases that might be chemicals. On-the-fly it has displayed the structure of nicotinic acid (which is not present anywhere in the original paper). OSCAR3 is acting as a "chemical amanuensis", who gives additional help readers who are not intimately familiar with the detailed science.
Data-mining in this form is rapid - OSCAR3 has been used to process half a million abstracts in a day. The benefits of running OSCAR over the scientific literature would be immense, yet the only text we can legally analyse is in few Open Source chemistry journals.
A major benefit comes when we can aggregate many different sources into one collection. It is not easy to do this with chemical publications because we are often forbidden to extract and aggregate data. However factual SI which is not protected by copyright gives an opportunity and we have done this for crystallographic data in our CrystalEye system.
The crystallographic community has for many years seen the value of re-use of factual data and was one of the first to advocate the publication of experimental data in full detail. Initially this was through paper copies sent to journals, which either archived them or forwarded them to the Cambridge Crystallographic Data Centre - a non-profit organization which abstracts crystallographic data from the literature, cleans and repackages it, and makes the aggregate available for an annual subscription (either nationally or per-laboratory). Many hundreds if not thousands of papers have been written on the results of data-mining the crystallography in this resource.
Over several decades the International Union of Crystallography has promoted the use of standard syntaxes and dictionaries for the publication of crystallography, culminating in the Crystallographic Information File (CIF) standard. It is now standard for all instruments to emit CIFs and for these to be used for scientific publication, sometimes as SI, sometimes incorporating the full-text. Several publishers, therefore, require that authors' CIFs are exposed as SI for peer-reviewed publications, and to make them Openly visible even if the full-text is closed.
Nick Day in our group has written software which harvests and aggregates these CIFs and converts the to XML (Chemical Markup Language, CML). He has found over 100,000 structures published in this way over the last 15 years. These are all hypopublished (i.e. no paper is explicitly coordinated with any other) but they are now aggregated on our site. Where the publisher makes it clear that the SI are not copyrighted we retain that; where the publisher claims copyright we have extracted the data and, even though we dispute their right, have not mounted the CIF.
In essence CrystalEye transforms the scholarly publication of crystallography into a giant knowledgebase of much greater power than the isolated articles. As an example the distance between a pair of atoms of specified types (for example, boron(B) and oxygen(O)) depends on the chemical environment. CrystalEye automatically plots the distribution of lengths (remote link on UCC site) :
| clickable diagram |
(This is an interactive SVG diagram which should display in all modern browsers and clicking on any bar will take you to the corresponding entries on the UCC site). If you cannot see the image, here is a non-interactive image
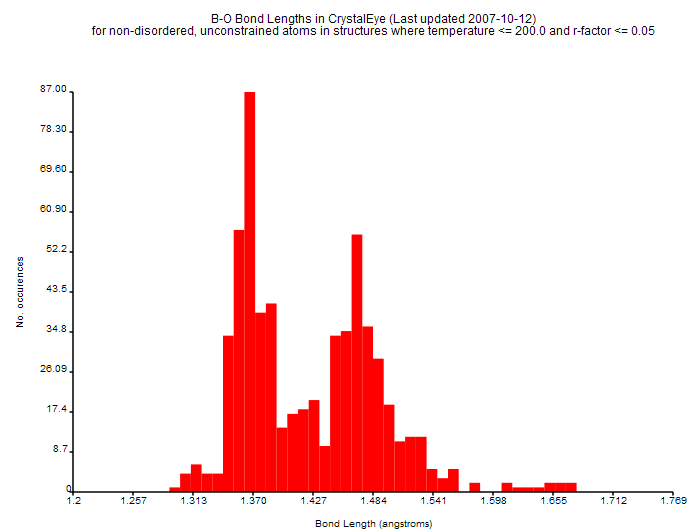 |
Only one Elsevier journal, Polyhedron, exposes its CIFs and is aggregated by crystaleye. I wrote to the editors of Tetrahedron (the sister journal for organic chemistry) six months ago requesting that they make their CIFs available for aggregation and I am still waiting for an acknowledgement.
Open data is young and the issues are not fully appreciated. A generous approach is to assume that most of the scholarly community do not yet realise the importance of Open Data. Recent initiatives such as the JISC/NSF report on cyberscholarship have emphasized the critical importance of data-driven scholarship. Within the cyber-community it is universally accepted that price and permission barriers to re-use of data are an enormous hurdle for cyberscholarship. We can expect that over the next decade successful examples of cyberscholarship will create major advocacy for Open Data.
The simplest and one of the most productive ways forward would be for the scholarly publishing community to take to heart the recommendations of the STM publishers and to enable Open Data in their products. This is, effectively done already by Open Access publishers who are fully BBB-compliant (e.g. have removed permission barriers) and who make this clear either by rubric or attaching an appropriate licence such as CC-BY. There are a number of "Open Access" publishers who do not explicitly remove permission barriers, or who offer a restricted product (most commonly "no commercial use"). It would be reasonable to expect that many of these did not realize the advantage to the community of converting to full permissionFree products and would then take this useful step.
At the other end of the spectrum there are closed-access publishers who nonetheless expose their SI as Open Data (the Royal Society of Chemistry is an example). This is a useful contribution, but it does not address the concern that much of the material in the full-text is factual and hence should be regarded as Open Data. Much of this stems from the continuation of paper journal publishing where the scope and extent of full-text was mandated by physical restrictions. There is no technical reason now why full-text and SI should be separated and why the complete scientific record should not be the object of publication as datuments. We predict that the demands of and vision of cyberscholarship will have a major influence in changing the traditional abbreviated full-text human-only document into a semantic machine-and-human-understandable hyperresource. Until that happens, however, it is important to insist that data are libre, whether embedded in full-text or elsewhere.
It is unlikely that even if they realise the value of Open Data all publishers will move rapidly towards the librefication of data. Many will feel that not only are their conventional publication models threatened by Open Access but those with profitable commercial database offerings will try to protect them by opposing Open Data. After all if all published data are Open, then what role is there for conventional databases?
This is a short-sighted view, because the new information models of "Web 2.0" are generating vast new businesses. There is no reason why freely available resources based on Open content should not generate new revenue streams. Risks will have to be taken, but the old model of generating revenue by limiting access to content which was originally free will come under increasing strain.
The success of the Open Source and Free Software movements has been due not only to advocacy but a large amount of attention to formalising the practice and subsequently monitoring and enforcing it. Initially the movements issued declarations of principles, such as Richard Stallman's four freedoms [1988]
These were rapidly followed up by formal licences such as the GNU General Public Licence [1989] which is a legal agreement derived from the freedoms and placing obligations on the author and user. In similar manner the Open Source Initiative [1998] enunciated similar principles and also maintains a list of licences compatible with these principles. Although there are critics of the terminology and many detailed wrinkles, "Open Source licence" is a sufficiently clear term to the vast majority of software developers and users that they do not need to inquire further before knowing what they can and cannot do.
By contrast the Open Access movement has only now started to realise the value of precise definitions of "Open Access" and to see to what extent those claiming to have OA products conformed to the BBB declarations. Consequently, although many publishers have clearly produced no-quibble OA products, there are many intermediate approaches which fail to comply with the full BBB declaration (permissionFree as well as priceFree). I have spent a considerable time trying to elicit the actual permission and restrictions on Open Access and Hybrid papers and the effort is substantial. Many do not have clear licences and use unstructured language spread over several web pages to indicate what is and is not expected.
In addition many publishers are effectively unhelpful in trying to help resolve these uncertainties and it is difficult to avoid regarding some of this as deliberate. I have written to several publishers over the last year about their policies and at least half have not bothered to reply and there is no contact address or number for general help. A typical example is given by Antony Williams who wishes to use our data transformed from the ACS's SI. On application he was told that he would have to wait at least 5 months for a discussion of the topic. As mentioned above, Elsevier's Tetrahedron has also not replied to me. It is not surprising, therefore, that publishers are often seen as an obstacle to Open Data rather than a solution.
It is a sign of progress that committed Open Access publishers have generally adopted CC licences as useful instruments. Full BBB-compliance requires CC-BY or CC-SA, so that CC-NC (non-commercial) falls short. However several publishers label themselves as Open Access but offer only CC-NC.
This fuzziness of labelling is a clear problem in Open Access where there are differences of view on permissionFreedom. Some, such as Stevan Harnad, contend that the fact of making a document publicly visible ("Green OA") is all that is necessary to remove permission barriers. Others such as Peter Suber and myself see this as insufficient since there can be no guarantee that the re-use of such material will not result in legal action. For that reason I believe that OA will only become mature when there are clear licences and labels for each publisher's offering.
The Open Data movement should therefore not rely solely on the progress of Open Access although this will be generally helpful in raising awareness of libre issues in general. I welcome the appearance of several groups in this area:The Open Knowledge Foundation was founded in 2004 "with the simple aim of promoting (and protecting) open knowledge in the belief that more open approaches to the production and distribution of knowledge have far-reaching social and commercial benefits. By 'open' knowledge we mean knowledge which anyone is free to use, re-use and redistribute without legal, social or technological restriction". The OKF have provided an OK definition to which works must conform:
1. Access. The work shall be available as a whole and at no more than a reasonable reproduction cost, preferably downloading via the Internet without charge. The work must also be available in a convenient and modifiable form.
2. Redistribution. The license shall not restrict any party from selling or giving away the work either on its own or as part of a package made from works from many different sources. The license shall not require a royalty or other fee for such sale or distribution.
3. Reuse. The license must allow for modifications and derivative works and must allow them to be distributed under the terms of the original work. The license may impose some form of attribution and integrity requirements: see principle 5 (Attribution) and principle 6 (Integrity) below.
4. Absence of Technological Restriction. The work must be provided in such a form that there are no technological obstacles to the performance of the above activities. This can be achieved by the provision of the work in an open data format, i.e. one whose specification is publicly and freely available and which places no restrictions monetary or otherwise upon its use.
5. Attribution.The license may require as a condition for redistribution and re-use the attribution of the contributors and creators to the work. If this condition is imposed it must not be onerous. For example if attribution is required a list of those requiring attribution should accompany the work.
6. Integrity. The license may require as a condition for the work being distributed in modified form that the resulting work carry a different name or version number from the original work.
7. No Discrimination Against Persons or Groups. The license must not discriminate against any person or group of persons.
8. No Discrimination Against Fields of Endeavor. The license must not restrict anyone from making use of the work in a specific field of endeavor. For example, it may not restrict the work from being used in a business, or from being used for military research.
9. Distribution of License. The rights attached to the work must apply to all to whom the work is redistributed without the need for execution of an additional license by those parties.
10. License Must Not Be Specific to a Package. The rights attached to the work must not depend on the work being part of a particular package. If the work is extracted from that package and used or distributed within the terms of the work's license, all parties to whom the work is redistributed should have the same rights as those that are granted in conjunction with the original package.
11. License Must Not Restrict the Distribution of Other Works. The license must not place restrictions on other works that are distributed along with the licensed work. For example, the license must not insist that all other works distributed on the same medium are open.
The OKF also maintains a list of licences compatible with the OKD, currently (2008-01) these include:
Open Data licences should be conformant to this set of meta-licence principles. There will be technical difficulties (that do not apply to OA) such as the complexity of data sets, the difficulty of downloading very large collections, the need to hold data in specialised engines, etc. Principle 4, therefore, may be de facto difficult to comply with and compliance may be judged by willingness to help. Note also that while an OA PDF requires no support a large compound data object, perhaps in RDF or XML, may be difficult to navigate and install. There is no obligation on the data provider to provide help free of charge.
The Talis Community Licence deserves special mention. Paul Miller writes
Creative Commons licenses are an extension of copyright law, as enshrined in the legal frameworks of various jurisdictions internationally. As such, it doesn't really work terribly well for a lot of (scientific, business, whatever) data... but the absence of anything better has led people to try slapping Creative Commons licenses of various types on data that they wish to share. ... Despite interest in open (or 'linked') data, licenses to provide protection (and, of course, to explicitly encourage reuse) are few and far between ... Building upon our original work on the TCL, we recently provided funding to lawyers Jordan Hatcher and Charlotte Waelde. They were tasked with validating the principles behind the license, developing an effective expression of those principles that could be applied beyond the database-aware shores of Europe, and working with us to identify a suitable home in which this new licence could be hosted, nurtured, and carried forward for the benefit of stakeholders far outside Talis. ...
Very recently [2007-12-17] Talis and Creative Commons (through its Science Commons project) announced that they were combining their efforts in the Open Data Commons Public Domain Dedication and Licence...
The Open Data Commons Public Domain Dedication & Licence is a document intended to allow you to freely share, modify, and use this work for any purpose and without any restrictions. This licence is intended for use on databases or their contents (”data”), either together or individually.
Many databases are covered by copyright. Some jurisdictions, mainly in Europe, have specific special rights that cover databases called the “sui generis” database right. Both of these sets of rights, as well as other legal rights used to protect databases and data, can create uncertainty or practical difficulty for those wishing to share databases and their underlying data but retain a limited amount of rights under a “some rights reserved” approach to licensing. As a result, this waiver and licence tries to the fullest extent possible to eliminate or fully license any rights that cover this database and data. Any Community Norms or similar statements of use of the database or data do not form a part of this document, and do not act as a contract for access or other terms of use for the database or data.
Science Commons has also developed a Protocol for Implementing Open Access Data. This is a meta-licence, allowing for the creation of conformant licences:
The Protocol is a method for ensuring that scientific databases can be legally integrated with one another. The Protocol is built on the public domain status of data in many countries (including the United States) and provides legal certainty to both data deposit and data use. The protocol is not a license or legal tool in itself, but instead a methodology for a) creating such legal tools and b) marking data already in the public domain for machine-assisted discovery.
The Open Data Commons Public Domain Dedication and License - the first legal tool to fully implement the Protocol. This draft is remarkable not just for the Public Domain Dedication but for the encoding of scholarly and scientific norms into a standalone, non-legal document. This is a key element of the Protocol and a major milestone in the fight for Open Access data.
There are two other developments from Creative Commons: CC0, a protocol that enables people to
(a) ASSERT that a work has no legal restrictions attached to it, OR
(b) WAIVE any rights associated with a work so it has not legal restrictions attached to it, and
(c) "SIGN" the assertion or waiver.
CC0 is similar to what the CC public domain dedication does now. The key addition is that the assertion that content is in the public domain will be vouched for by users, so that there is a platform for reputation systems to develop. People will then be able to judge the reliability of content's copyright status based on who has done the certifying.
and Community Norms, which has no legal implications but acts as an exhortation to develop communal collaborative best practice.
You are free to follow or ignore all or part of the norms listed below. The below text however reflects norms of access and use that can help create a vibrant community around data and databases that would be great if you would voluntarily adopt.
We, the user and provider community of this database and data, wish to declare that these are the norms of our community. This is our code of conduct; our “best practices” guide and we would like to see members of our community participate in the following ways:
To help clarify this John Wilbank of Science Commons wrote:
The way we at Science Commons have tried to frame this issue is that the best method is to converge on the public domain. Thus, the way to evaluate any one license or terms of use is to see if they waive rights, not reserve them and use them. Please see the protocol at http://sciencecommons.org/projects/publishing/open-access-data-protocol/ and a blog post at http://sciencecommons.org/weblog/archives/2007/12/16/announcing-protocol-for-oa-data/for more information. Note that all ideas like attribution and share-alike are disallowed as legal constraints under the protocol, but encouraged under "norms" documents.
The only "license" or "legal tool" that implements the protocol is the Public Domain Dedication tool that Talis sponsored and Jordan wrote. CCZero will comply from a legal perspective, and we will encourage everyone to simply use the norms guidance that Jordan wrote as well rather than replicate that work. CC will provide metatdata about the legal status of the public domain, and the PDDL will be certified to use that metadata and all trademarks. SC will provide additional metadata and marks around the norms Jordan wrote.
The reason for the protocol is that, rather than trying to force everyone to converge around a single license - especially given how much data is already in the public domain - we prefer to "mark" databases as open using certified terms of use or licenses. This allows a lot of different implementations of the protocol. The NCBI terms of use at http://www.ncbi.nlm.nih.gov/About/disclaimer.html are a good example of a pre-existing set of terms we want to certify as compliant.
The next step is a "mark the data world" project in which users and database providers make assertions of openness using the metadata, either through RDFa or true RDF or microformats. That will be followed by the integration of the metadata into search engines so that one can search for public domain compliant data for all forms of reuse, but also easily find the norms associated.
The other key here is what one's goals are. In the case of scientific data and data integration, we at SC are convinced that it's imperative to keep share alike and attribution outside the legal document. I know that Rufus [Pollock, founder of OKF] disagrees with this conclusion in the case of non-science data, and it's a matter on which reasonable people can disagree.
These developments are very recent and have taken a great deal of hard work so it is difficult to describe them accurately in this review. However it is clear that they are a major achievement and are likely to form the basis for Open Data for the foreseeable future. They represent a great degree of community agreement, so that there are no obvious competing approaches ("We think it’s important to avoid legal fragmentation at the early stages, and that one way to avoid that fragmentation is to work with the existing thought leaders like the OKF."). They make clear that data are different from manuscripts. Manuscripts can be copyrighted whereas facts are in the public domain. The protection of the public domain is difficult and has required considerable legal input.
Open Data in science is now recognised as a critically important area which needs much careful and coordinated work if it is to develop successfully. Much of this requires advocacy and it is likely that when scientists are made aware of the value of labeling their work the movement will grow rapidly. Besides the licences and buttons there are other tools which can make it easier to create Open Data (for example modifying software so that it can mark the work and also to add hash codes to protect the digital integrity).
Creative Commons is well known outside Open Access and has a large following. Outside of software, it is seen by many as the default way of protecting their work while making it available in the way they wish. CC has the resources, the community respect and the commitment to continue to develop appropriate tools and strategies.
But there is much more that needs to be done. Full Open Access is the simplest solution but if we have to coexist with closed full-text the problem of embedded data must be addressed, by recognising the right to extract and index data. And in any case conventional publication discourages the full publication of the scientific record. The adoption of Open Notebook Science in parallel with the formal publications of the work can do much to liberate the data. Although data quality and formats are not strictly part of Open Data, its adoption will have parked improvements. The general realisation of the value of reuse will create strong pressure for more and better data. If publishers do not gladly accept this challenge, then scientists will rapidly find other ways of publishing data, probably through institutional, departmental, national or international subject repositories. In any case the community will rapidly move to Open Data and publishers resisting this will be seen as a problem to be circumvented.